
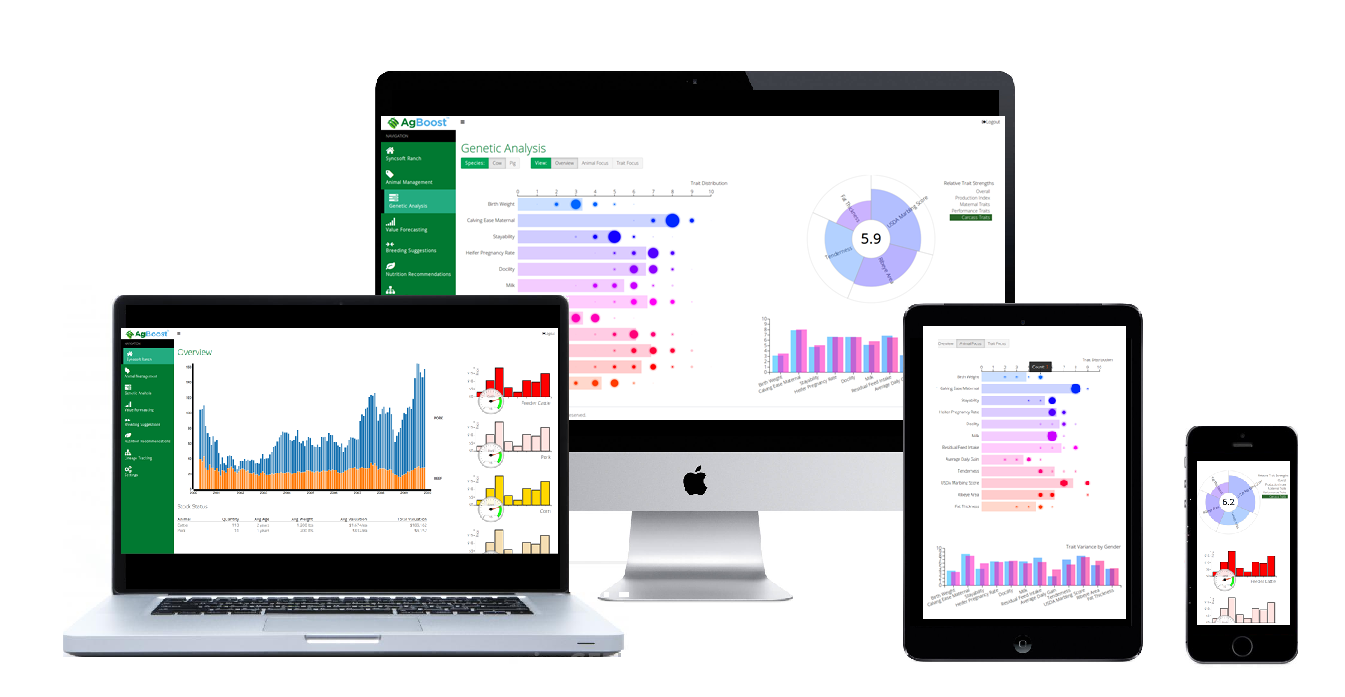
Supports DNA and Protein multiple sequence alignments via Clustal And Muscles plugins. Design cloning strategies, design primers, and create beautiful plasmid maps that can be edited and adjusted any way you want. Assemble sequencing data, analyse mutations, and export the results. Import or retrieve annotated sequence files from a variety of formats and online databases. Additionally, affection status alone can be used to construct useful segregation analysis of complex family pedigrees to find candidate regions and variants.DNADynamo DNA sequencing and Analysis Software is easy to use. Custom filters can be easily created using family information. Prebuilt filters are available for most common inheritance patterns. Inheritance and Affection Status Based Filtering: VarSeq includes first-class support for workflows that use pedigree, affection status, or custom groupings to define filter criteria.Additional statistics, such as distance from coding start, distance from exon boundary, and the number of codons changed make it easy to examine specific classes of variants. Each variant is mapped to all overlapping transcripts and information about the region where it is located (exon, intron, intergenic, etc.), sequence ontology (frame shift, synonymous, etc.), and HGVS notation (g dot, c dot, and p dot) is provided. Powerful Variant Annotation: Included in VarSeq is functionality similar to SnpEff or Variant Effect Predictor.These generated rankings can be combined with traditional filtering techniques to quickly surface candidate variants. By examining biomedical ontologies that link diseases with genes, the algorithm can rank genes according to the proband's phenotype.
 Phenotype Based Gene Ranking: Variants can be prioritized by examining how closely the mutated gene is related to the patient's phenotype. This includes using related samples in trios and quad analysis, algorithms that detect inheritance patterns of variants, ranking of rare patheogenic variants by their relevance to the phenotype terms provided by differential diagnositics and the integration of the public and premium annotations sources. Rare Disease Diagnosis: VarSeq includes first-class support for the more complex workflows associated with discovering the causal variants of rare disease. Supported Genetics Tests Include: Diagnostic testing used to identify the presence or absence of causal genetic variations for specific disease, predictive and pre-symptomatic genetics tests designed to identify hereditary gene changes that can increase the lifetime risk of developing diseases, and Newborn Screening used to test babies soon after birth to identify highly penetrant variants for certain diseases known to cause problems with health and development. VarSeq's flexibility allows you to customize workflows to the individual gene panel or exome test. Comprehensive Workflows: The VarSeq clinical stack supports all of the steps necessary to provide clinical genetic tests from the raw VCF variants to the signed out clinical report. As with all of our public annotation libraries, COSMIC is updated regularly, providing you with the most recent and relevant data. COSMIC breaks out each sample-variant pair into a record, and VarSeq provides the fields in COSMIC with relevant hyperlinks and formatting to enable easy filtering. VarSeq also provides the gene level annotations with relevant summary and curated oncology details provided by COSMIC. The release of COSMIC V71 integrated The Cancer Genome Atlas, making it the most compressive catalog of somatic mutations in history.
Phenotype Based Gene Ranking: Variants can be prioritized by examining how closely the mutated gene is related to the patient's phenotype. This includes using related samples in trios and quad analysis, algorithms that detect inheritance patterns of variants, ranking of rare patheogenic variants by their relevance to the phenotype terms provided by differential diagnositics and the integration of the public and premium annotations sources. Rare Disease Diagnosis: VarSeq includes first-class support for the more complex workflows associated with discovering the causal variants of rare disease. Supported Genetics Tests Include: Diagnostic testing used to identify the presence or absence of causal genetic variations for specific disease, predictive and pre-symptomatic genetics tests designed to identify hereditary gene changes that can increase the lifetime risk of developing diseases, and Newborn Screening used to test babies soon after birth to identify highly penetrant variants for certain diseases known to cause problems with health and development. VarSeq's flexibility allows you to customize workflows to the individual gene panel or exome test. Comprehensive Workflows: The VarSeq clinical stack supports all of the steps necessary to provide clinical genetic tests from the raw VCF variants to the signed out clinical report. As with all of our public annotation libraries, COSMIC is updated regularly, providing you with the most recent and relevant data. COSMIC breaks out each sample-variant pair into a record, and VarSeq provides the fields in COSMIC with relevant hyperlinks and formatting to enable easy filtering. VarSeq also provides the gene level annotations with relevant summary and curated oncology details provided by COSMIC. The release of COSMIC V71 integrated The Cancer Genome Atlas, making it the most compressive catalog of somatic mutations in history. 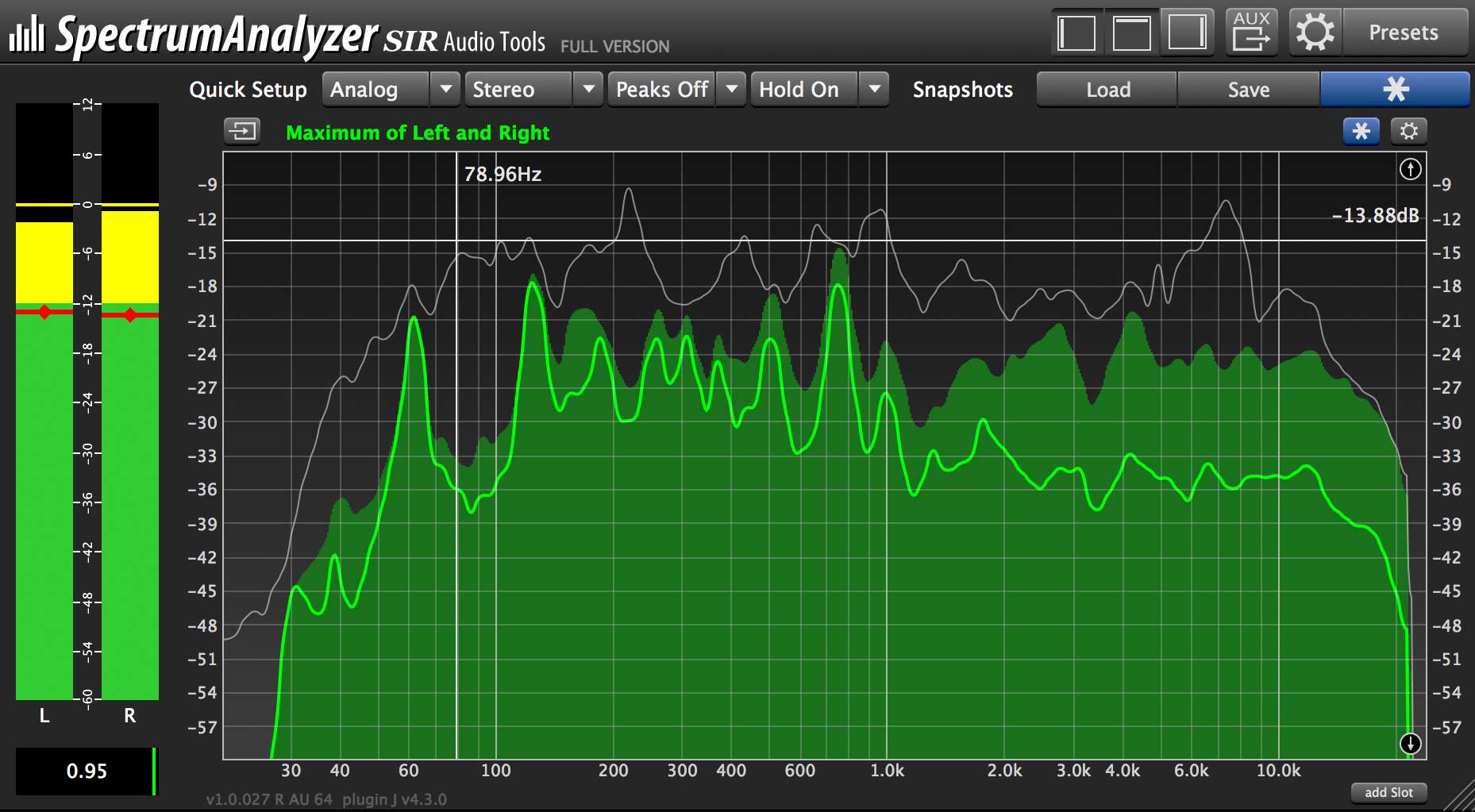
COSMIC is a population catalog containing somatic mutations related to human cancer.
COSMIC: Included in VarSeq is the Catalogue of Somatic Mutations in Cancer or COSMIC. Multiple paired samples can be imported in a single project enabling fast and accurate analysis in settings where reproducibility is critical. Samples can be imported as matched pairs, allowing germ line variants to be filtered out in a single step. 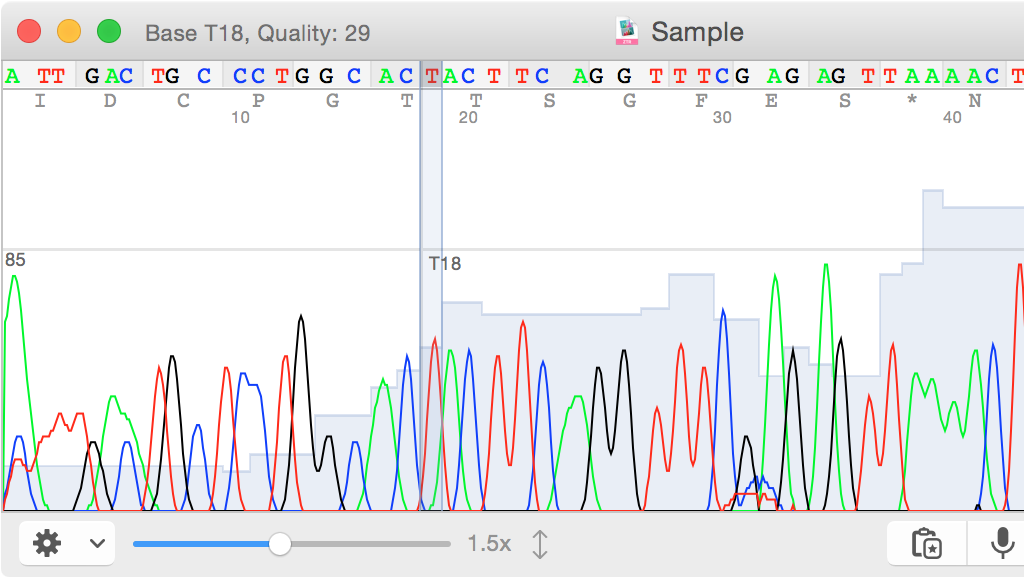
Tumor/Normal Workflows: VarSeq provides complete support for Tumor Normal workflows. Whole Exome and Whole Genome Analysis: VarSeq enables cutting edge cancer analysis by easily supporting whole exome and genome sized datasets. Finally, variant blacklists can be imported to filter variants that your lab has found to be inaccurate or not actionable. Additionally, variants can be annotated precisely against COSMIC, fully utilizing the many somatic mutation annotations present in the database. Allelic ratios are automatically calculated to detect somatic mutations at low tumor fractions. Support for Cancer Gene Panels: VarSeq includes the specialized features needed for cancer workflows.








 0 kommentar(er)
0 kommentar(er)
